- Inhibitors
- Antibodies
- Compound Libraries
- New Products
- Contact Us
research use only
MS023 Histone Methyltransferase inhibitor
Cat.No.S8112
Chemical Structure
Molecular Weight: 287.40
Jump to
Quality Control
Batch:
Purity:
99.15%
99.15
| Related Targets | HDAC JAK BET PKC PARP HIF PRMT EZH2 AMPK Histone Acetyltransferase |
|---|---|
| Other Histone Methyltransferase Inhibitors | Pinometostat (EPZ5676) 3-Deazaneplanocin A (DZNep) Hydrochloride BIX-01294 trihydrochloride EPZ004777 EPZ015666 (GSK3235025) UNC1999 EPZ005687 SGC 0946 MM-102 UNC0638 |
Solubility
|
In vitro |
DMSO
: 57 mg/mL
(198.32 mM)
Ethanol : 57 mg/mL Water : Insoluble |
Molarity Calculator
Dilution Calculator
Molecular Weight Calculator
|
In vivo |
|||||
In vivo Formulation Calculator (Clear solution)
Step 1: Enter information below (Recommended: An additional animal making an allowance for loss during the experiment)
mg/kg
g
μL
Step 2: Enter the in vivo formulation (This is only the calculator, not formulation. Please contact us first if there is no in vivo formulation at the solubility Section.)
%
DMSO
%
%
Tween 80
%
ddH2O
%
DMSO
+
%
Calculation results:
Working concentration: mg/ml;
Method for preparing DMSO master liquid: mg drug pre-dissolved in μL DMSO ( Master liquid concentration mg/mL, Please contact us first if the concentration exceeds the DMSO solubility of the batch of drug. )
Method for preparing in vivo formulation: Take μL DMSO master liquid, next addμL PEG300, mix and clarify, next addμL Tween 80, mix and clarify, next add μL ddH2O, mix and clarify.
Method for preparing in vivo formulation: Take μL DMSO master liquid, next add μL Corn oil, mix and clarify.
Note: 1. Please make sure the liquid is clear before adding the next solvent.
2. Be sure to add the solvent(s) in order. You must ensure that the solution obtained, in the previous addition, is a clear solution before proceeding to add the next solvent. Physical methods such
as vortex, ultrasound or hot water bath can be used to aid dissolving.
Chemical Information, Storage & Stability
| Molecular Weight | 287.40 | Formula | C17H25N3O |
Storage (From the date of receipt) | |
|---|---|---|---|---|---|
| CAS No. | 1831110-54-3 | Download SDF | Storage of Stock Solutions |
|
|
| Synonyms | N/A | Smiles | CC(C)OC1=CC=C(C=C1)C2=CNC=C2CN(C)CCN | ||
Mechanism of Action
| Targets/IC50/Ki |
PRMT6
(Cell-free assay) 4 nM
PRMT8
(Cell-free assay) 5 nM
PRMT1
(Cell-free assay) 30 nM
PRMT4
(Cell-free assay) 83 nM
PRMT3
(Cell-free assay) 119 nM
|
|---|---|
| In vitro |
MS023 potently reduces cellular levels of H4R3me2a in MCF7 and HEK293 cells by inhibiting PRMT1/6 methyltransferase activity with IC50 of 9 nM and 56 nM, respectively. This compound also inhibits cell growth and potentially induces growth arrest and flattening morphology at low concentrations. It displayed high potency for type I PRMTs including PRMT1, 3, 4, 6 and 8, but was completely inactive against type II and type III PRMTs, protein lysine methyltransferases and DNA methyltransferases. This chemical potently decreased cellular levels of histone arginine asymmetric dimethylation. It also reduced global levels of arginine asymmetric dimethylation and concurrently increased levels of arginine monomethylation and symmetric dimethylation in cells. |
| Kinase Assay |
PRMT Biochemical Assays
|
|
A scintillation proximity assay (SPA) is used for assessing the effect of test compounds on inhibiting the methyl transfer reaction catalyzed by PRMTs. In brief, the tritiated S-adenosyl-L-methionine (3H-SAM) is used as the donor of methyl group. The (3H) methylated biotin labeled peptide is captured in a streptavidin/scintillant-coated microplate, which brings the incorporated 3H-methyl and the scintillant to close proximity resulting in light emission that is quantified by tracing the radioactivity signal (counts per minute) as measured by a TopCount NXT Microplate Scintillation and Luminescence Counter. When necessary, nontritiated SAM is used to supplement the reactions. The IC50 values are determined under balanced conditions at Km concentrations of both substrate and cofactor by titration of test compounds in the reaction mixture.
|
|
| In vivo |
This compound is a potent, selective, and cell-active Type I PRMT inhibitor. |
References |
|
Applications
| Methods | Biomarkers | Images | PMID |
|---|---|---|---|
| Western blot | PRMT6 / H3R2me2a / DNMT1 / UHRF1 |
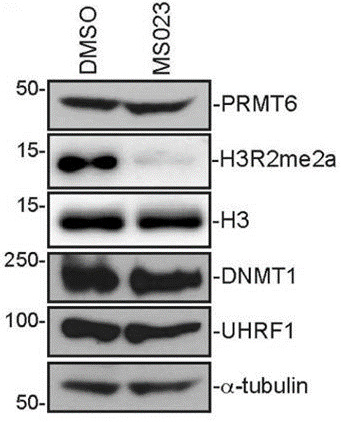
|
29262320 |
Tech Support
Tel: +1-832-582-8158 Ext:3
If you have any other enquiries, please leave a message.






































