|
Toll Free: (877) 796-6397 -- USA and Canada only -- |
Fax: +1-832-582-8590 Orders: +1-832-582-8158 |
Tech Support: +1-832-582-8158 Ext:3 Please provide your Order Number in the email. |
Technical Data
| Formula | C22H24N4O4 |
||||||
| Molecular Weight | 408.45 | CAS No. | 1173900-33-8 | ||||
| Solubility (25°C)* | In vitro | DMSO | 82 mg/mL (200.75 mM) | ||||
| Ethanol | 10 mg/mL (24.48 mM) | ||||||
| Water | Insoluble | ||||||
| In vivo (Add solvents to the product individually and in order) |
|
||||||
|
* <1 mg/ml means slightly soluble or insoluble. * Please note that Selleck tests the solubility of all compounds in-house, and the actual solubility may differ slightly from published values. This is normal and is due to slight batch-to-batch variations. * Room temperature shipping (Stability testing shows this product can be shipped without any cooling measures.) |
|||||||
Preparing Stock Solutions
Biological Activity
| Description | AZD6482 (KIN-193) is a PI3Kβ inhibitor with IC50 of 10 nM, 8-, 87- and 109-fold more selective to PI3Kβ than PI3Kδ, PI3Kα and PI3Kγ in cell-free assays. Phase 1. | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Targets |
|
||||||||
| In vitro | AZD6482 also inhibits PI3Kα, γ, and δ, with IC50 of 80 nM to 1.09 μM, which are significantly lower than its (+)-enantiomer (S-form). This compound is an antiplatelet agent; it blocks platelet activation adhesion/aggregation and promotes platelet disaggregation in assay of washed platelet aggregation (WPA), with an IC50 value of 6 nM. Furthermore, by targeting PI3Kβ, this chemical specifically inhibits thrombosis without interfering with normal haemostasis. Therefore, it is used as an anti-thrombotic drug for the prophylaxis of thrombotic disorders. [1] |
Protocol (from reference)
| Kinase Assay:[1] |
|
|---|---|
| Cell Assay:[1] |
|
References
|
Customer Product Validation
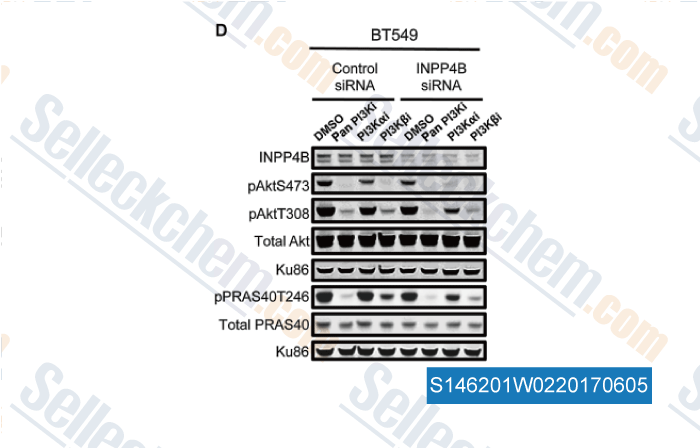
-
, , Mol Cancer Res, 2017, 15(6):765-775

-
, , One customer
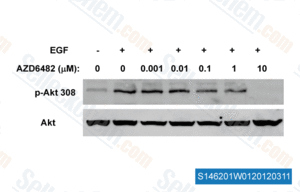
-
, , Dr. Zhang of Tianjin Medical University
![Collagen-embedded melanoma spheroids were treated for 72 h with SGI-1776 [5 μM] or AZD6482 [10 μM] as single agents or in combination before staining for live (green) and dead (red) cells. Experiments were conducted in triplicate and representative images are shown. Scale bar represents 150 microns.](https://file.selleckchem.com/downloads/review/700px/AZD6482-S146201Y0320170605.gif)
-
Data from [ , , Oncotarget, 2016, 7(34):54897-54912 ]
Selleck's AZD6482 Has Been Cited by 28 Publications
| Human colonic organoids for understanding early events of familial adenomatous polyposis pathogenesis [ J Pathol, 2025, 265(1):26-40] | PubMed: 39641466 |
| Longitudinal phosphoproteomics reveals the PI3K-PAK1 axis as a potential target for recurrent colorectal liver metastases [ Cell Rep, 2024, 43(12):115061] | PubMed: 39689713 |
| Molecular mechanisms of PI3K isoform dependence in embryonic growth [ J Turk Ger Gynecol Assoc, 2024, 25(3):159-166] | PubMed: 39219229 |
| A Gene Co-Expression Network-Based Drug Repositioning Approach Identifies Candidates for Treatment of Hepatocellular Carcinoma [ Cancers (Basel), 2022, 14(6)1573] | PubMed: 35326724 |
| ZAP70 Activation Compensates for Loss of Class IA PI3K Isoforms Through Activation of the JAK-STAT3 Pathway [ Cancer Diagn Progn, 2022, 2(3):391-404] | PubMed: 35530641 |
| Synergism between the phosphatidylinositol 3-kinase p110β isoform inhibitor AZD6482 and the mixed lineage kinase 3 inhibitor URMC-099 on the blockade of glioblastoma cell motility and focal adhesion formation [ Cancer Cell Int, 2021, 21(1):24] | PubMed: 33407478 |
| Metabolic Imaging Detects Resistance to PI3Kα Inhibition Mediated by Persistent FOXM1 Expression in ER+ Breast Cancer [ Cancer Cell, 2020, S1535-6108(20)30427-X] | PubMed: 32976773 |
| PIK3CA C-terminal frameshift mutations are novel oncogenic events that sensitize tumors to PI3K-α inhibition [ Proc Natl Acad Sci U S A, 2020, 117(39):24427-24433] | PubMed: 32929011 |
| Targeting phosphatidylinositol 3 kinase-β and -δ for Bruton tyrosine kinase resistance in diffuse large B-cell lymphoma [ Blood Adv, 2020, 4(18):4382-4392] | PubMed: 32926124 |
| Application of a Biphasic Mathematical Model of Cancer Cell Drug Response for Formulating Potent and Synergistic Targeted Drug Combinations to Triple Negative Breast Cancer Cells. [ Cancers (Basel), 2020, 27;12(5)pii: E1087] | PubMed: 32349331 |
RETURN POLICY
Selleck Chemical’s Unconditional Return Policy ensures a smooth online shopping experience for our customers. If you are in any way unsatisfied with your purchase, you may return any item(s) within 7 days of receiving it. In the event of product quality issues, either protocol related or product related problems, you may return any item(s) within 365 days from the original purchase date. Please follow the instructions below when returning products.
SHIPPING AND STORAGE
Selleck products are transported at room temperature. If you receive the product at room temperature, please rest assured, the Selleck Quality Inspection Department has conducted experiments to verify that the normal temperature placement of one month will not affect the biological activity of powder products. After collecting, please store the product according to the requirements described in the datasheet. Most Selleck products are stable under the recommended conditions.
NOT FOR HUMAN, VETERINARY DIAGNOSTIC OR THERAPEUTIC USE.
