|
How to Cite 1. For In-Text Citation (Materials & Methods): 2. For Key Resources Table: |
||
|
Toll Free: (877) 796-6397 -- USA and Canada only -- |
Fax: +1-832-582-8590 Orders: +1-832-582-8158 |
Tech Support: +1-832-582-8158 Ext:3 Please provide your Order Number in the email. We strive to reply to |
Technical Data
| Formula | C29H32N4O3S |
||||||||||
| Molecular Weight | 516.65 | CAS No. | 422513-13-1 | ||||||||
| Solubility (25°C)* | In vitro | DMSO | 100 mg/mL (193.55 mM) | ||||||||
| Water | Insoluble | ||||||||||
| Ethanol | Insoluble | ||||||||||
| In vivo (Add solvents to the product individually and in order) |
|
||||||||||
|
* <1 mg/ml means slightly soluble or insoluble. * Please note that Selleck tests the solubility of all compounds in-house, and the actual solubility may differ slightly from published values. This is normal and is due to slight batch-to-batch variations. * Room temperature shipping (Stability testing shows this product can be shipped without any cooling measures.) |
|||||||||||
Preparing Stock Solutions
Biological Activity
| Description | Hesperadin potently inhibits Aurora B with IC50 of 250 nM in a cell-free assay. It markedly reduces the activity of AMPK, Lck, MKK1, MAPKAP-K1, CHK1 and PHK while this compound does not inhibit MKK1 activity in vivo. | ||||
|---|---|---|---|---|---|
| Targets |
|
||||
| In vitro | Hesperadin inhibits the ability of immunoprecipitated Aurora B to phosphorylate histone H3 with IC50 of 250 nM and markedly reduces the activity of other kinases (AMPK, Lck, MKK1, MAPKAP-K1, CHK1, and PHK) at a concentration of 1 μM. In contrast, only 20-100 nM of this compound is sufficient to induce the loss of mitotic histone H3-Ser10 phosphorylation in HeLa cells. Its treatment causes defects in mitosis and cytokinesis, leading to stoppage of proliferation of HeLa cells and polyploidization, which can be specifically ascribed to the inhibition of Aurora B function during the process of chromosome attachment. This compound (100 nM) quickly overrides the mitotic arrest induced by taxol or monastrol but not by nocodazole. It and nocodazole treatment in HeLa cells abolishes kinetochore localization of BubR1 and diminishes the intensity of Bub1 at kinetochores, suggesting that Aurora B function is required for efficient kinetochore recruitment of BubR1 and Bub1, which in turn might be necessary for prolonged checkpoint signaling. [1] It prevents the phosphorylation of recombinant trypanosome histone H3 by the T. brucei Aurora kinase-1 (TbAUK1) from pathogenic Trypanosoma brucei with IC50 of 40 nM in vitro kinase assays. This chemical significantly inhibits cell growth of cultured infectious bloodstream forms (BF) with IC50 of 48 nM, and only weakly inhibits cell growth of insect stage procyclic forms (PF) with IC50 of 550 nM. [2] |
Protocol (from reference)
| Kinase Assay:[1] |
|
|---|---|
| Cell Assay:[1] |
|
References
|
Customer Product Validation
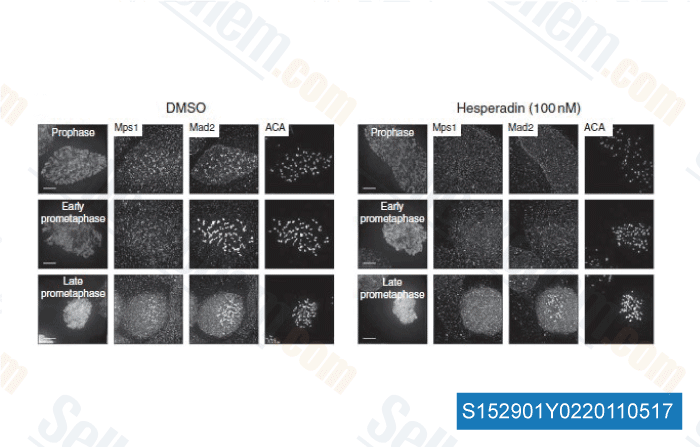
-
Data from [ Nat Commun , 2011 , 2, 316 ]
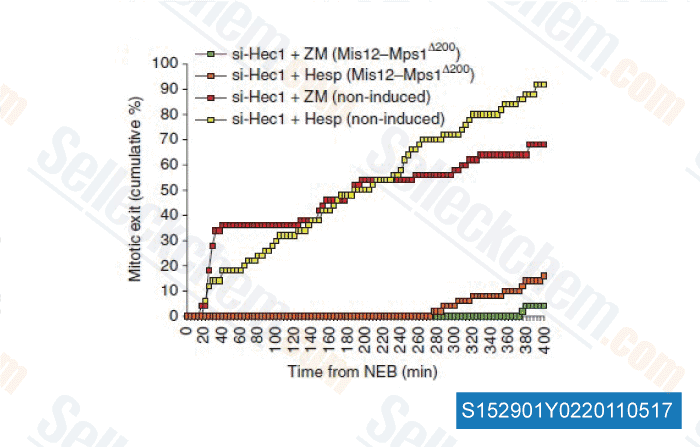
-
Data from [ Nat Commun , 2011 , 2, 316 ]
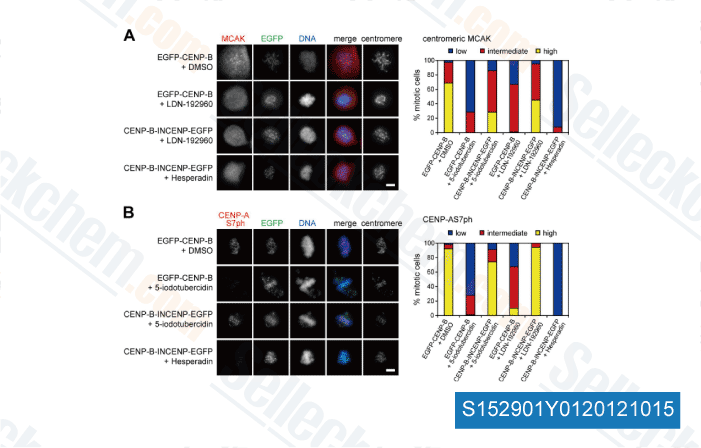
-
,
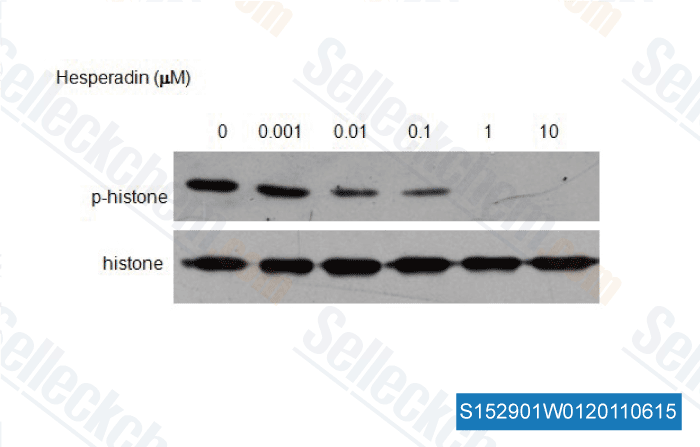
-
, , Dr. Zhang of Tianjin Medical University
Selleck's Hesperadin Has Been Cited by 51 Publications
| Homologous recombination promotes non-immunogenic mitotic cell death upon DNA damage [ Nat Cell Biol, 2025, 27(1):59-72] | PubMed: 39805921 |
| A CPC-shelterin-BTR axis regulates mitotic telomere deprotection [ Nat Commun, 2025, 16(1):2277] | PubMed: 40097392 |
| Identification of chemical inhibitors targeting long noncoding RNA through gene signature-based high throughput screening [ Int J Biol Macromol, 2025, 292:139119] | PubMed: 39722392 |
| Aurora B maintains spherical shape of mitotic cells via simultaneously stabilizing myosin II and vimentin [ J Mol Cell Biol, 2025, mjaf023] | PubMed: 40795355 |
| Dynamic phosphorylation of FOXA1 by Aurora B guides post-mitotic gene reactivation [ Cell Rep, 2024, 43(9):114739] | PubMed: 39276350 |
| Visualization strategies to aid interpretation of high-dimensional genotoxicity data [ Environ Mol Mutagen, 2024, 10.1002/em.22604] | PubMed: 38757760 |
| Histone H3 phospho-regulation by KimH3 in both interphase and mitosis [ iScience, 2023, 26(4):106372] | PubMed: 37013187 |
| Rsk2 and Lrp5 deficiency limit osteosarcoma growth in cFos-transgenic mice by different mechanisms [ ProQuest , 2023, 30683696] | PubMed: None |
| Hesperadin suppresses pancreatic cancer through ATF4/GADD45A axis at nanomolar concentrations [ Oncogene, 2022, 10.1038/s41388-022-02328-4] | PubMed: 35551503 |
| CDC20-Mediated hnRNPU Ubiquitination Regulates Chromatin Condensation and Anti-Cancer Drug Response [ Cancers (Basel), 2022, 14(15)3732] | PubMed: 35954396 |
RETURN POLICY
Selleck Chemical’s Unconditional Return Policy ensures a smooth online shopping experience for our customers. If you are in any way unsatisfied with your purchase, you may return any item(s) within 7 days of receiving it. In the event of product quality issues, either protocol related or product related problems, you may return any item(s) within 365 days from the original purchase date. Please follow the instructions below when returning products.
SHIPPING AND STORAGE
Selleck products are transported at room temperature. If you receive the product at room temperature, please rest assured, the Selleck Quality Inspection Department has conducted experiments to verify that the normal temperature placement of one month will not affect the biological activity of powder products. After collecting, please store the product according to the requirements described in the datasheet. Most Selleck products are stable under the recommended conditions.
NOT FOR HUMAN, VETERINARY DIAGNOSTIC OR THERAPEUTIC USE.
